Who says neuroscientists don’t need more brains? Annotation with SciBot
You might think that neuroscientists already have enough brains, but apparently not. Over 100 neuroscientists attending the recent annual meeting of the Society for Neuroscience (SFN), took part in an annotation challenge: modifying scientific papers to add simple references that automatically generate and attach Hypothesis annotations, filled with key related information. To sweeten the pot, our friends at Gigascience gave researchers who annotated their own papers their very own brain hats.
But handing out brains is not just a conference gimmick. Thanks to our colleagues at the Neuroscience Information Framework (NIF), Hypothesis was once again featured at SFN, the largest gathering of neuroscientists in the world, attended by well over 30,000 people in San Diego Nov 12-16, 2016. The annotation challenge at SFN was a demonstration of a much larger collaboration with NIF: to increase rigor and reproducibility in neuroscience by using the NIF’s new SciBot service to annotate publications automatically with links to related materials and tools that researchers use in scientific studies.
These research resources and how they are reported in scientific papers has been a concern of NIF since 2008 (see Bandrowski et al., 2015 for more details). As NIF and others have demonstrated, researchers aren’t always as thorough as they should be when reporting the materials and methods on which their findings rest. To improve reporting standards, NIF has spearheaded the effort to have authors supply a standard syntax that includes a unique identifier, called an RRID (research resource identifier) for specifying exactly which resources they used in a paper. The RRID standard is now used by more than 150 journals within biomedicine.
In our big splash at SFN, NIF and Hypothesis released a collection of thousands of RRID annotations generated by SciBot, our hybrid machine- and human-based annotation tool. When activated, SciBot automatically recognizes RRIDs within papers, then makes a call to an RRID resolver service and pipes the information about these resources—including other papers that have been published using them—into annotations managed by the Hypothesis client.
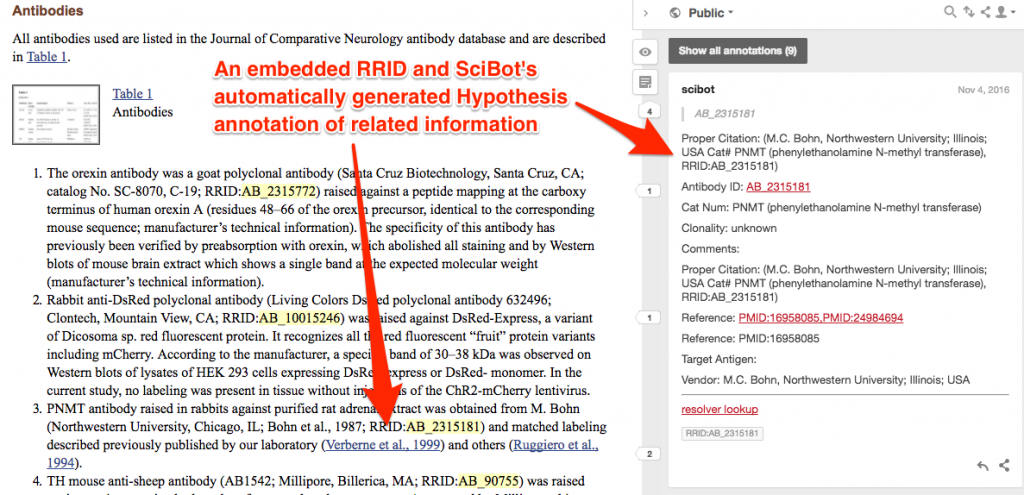
The NIF team initially used SciBot as a curation tool to see how well it recognized and resolved RRIDs—data that will be used to help tune SciBot’s recognition algorithm. The curators also analyzed how well authors were supplying and formatting RRIDs according to standards. Building on the success of the curation activity, we have now released a validated data set of SciBot’s annotations to the public Hypothesis channel.
After debuting Hypothesis last year at SFN 2015, we were pleased to see other reports of Hypothesis in use, like in the presentation by Dr. Elba Serrano at New Mexico State University who used annotation to help graduate students apply the new reproducibility and rigor guidelines issued by the US National Institutes of Health.
Authoritative Annotation
Back at the 2015 SFN meeting, neuroscientists were both excited and nervous about the possibilities of web-based annotation. Would it be used to improve communications or trash scientific reputations? We think SciBot’s annotations for resource identification demonstrated in 2016 highlight a powerful use of Hypothesis to enrich science. We are calling this innovation “authoritative annotation,” where an expert agent—human or otherwise—layers additional related data on top of scientific articles or other primary sources. We expect authoritative annotations to facilitate higher levels of rigor and reproducibility in biomedicine, particularly in the area of biocuration, and hope to support authoritative annotation in other science and research areas.